Ancient DNA: Methods and Protocols (8 page)
Read Ancient DNA: Methods and Protocols Online
Authors: Beth Shapiro
and collected a second elution of 100 m L in case any DNA remained on the column membrane. This extraction represents the sample from which the full 445 bp of nuclear DNA was generated
( 1
) .
Second, we extracted a small section of toe pad using the silica protocol for DNA extraction from bones and teeth described in Chap. 3 . Initially, we used in 10 mL of 0.5 M EDTA and proteinase K as per the protocol, incubating the sample at room temperature with rotation overnight. After 24 h, the toe pad remained largely intact, so we added an additional 5 mg of proteinase K and incubated the sample at room temperature with rotation for a further 24 h. As the toe pad still had not dissolved after this step, we then moved the solution to an incubator set at 50°C with rotation for a third 24 h period, then changed the temperature to 30°C for a fourth day of incubation with rotation. We added another 10 mg of proteinase K and continued to incubate the sample at 30°C with rotation for 4 more days. Finally, we added an additional 10 mg of proteinase K and incubated the sample with rotation for a fi nal 24 h at 50°C, which eventually yielded a completely dissolved toe pad. We then processed the fi nal solution following the silica extraction protocol.
We selected intron 7 of the nuclear-encoded
fi brinogen beta chain (FGB)
gene for sequencing. Due to the level of fragmentation incurred by aDNA, we designed a series of overlapping primer sets (Table 1 in
( 1
) ) to amplify fragments no longer than 170 bp, including the primers. To design the primers, we downloaded and aligned all available
FGB intron 7
sequences from GenBank. We used SeqBuilder v8.1 (DNASTAR) to design primers, which we selected to bind in regions that are conserved across the
Columbidae
.
When we could not identify conserved regions, primers were designed to match most closely previously sequenced species belonging to the typical pigeons and doves
( 6 )
. We compared the results of the two extraction protocols by attempting to amplify nuclear DNA using primer set
FGB
-F6R7.
32
T.L. Fulton
et al.
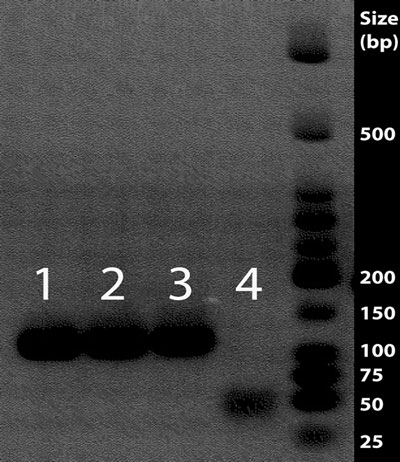
Fig. 1. Gel image of
FGB
F6R7 amplifi cation. PCR products are visualized with ethidium bromide and UV illumination. Each
lane
contains 5 m L of 25 m L reactions with templates from (
left
to
right
): Qiagen fi rst elution, Qiagen second elution, silica extraction, PCR negative, and 6 m L of a 50% dilution of NEB low molecular weight ladder. Primer–dimer can be observed in the PCR negative lane (
lane 4
), forming in the absence of template DNA.
We performed PCR amplifi cations in 25 m L reactions comprising 50 m g rabbit serum albumin, 0.25 mM dNTPs, 1× high fi delity buffer, 1.25 units Platinum
Taq
High Fidelity (Invitrogen), 3 mM
MgSO , 1 m M of each primer, and 1 m L DNA extract. Cycling 4
conditions were 94°C for 90 s, 60 cycles of 94°C for 45 s, 45 s at 48°C, 68°C for 90 s followed by 10 min of 68°C. We used a negative PCR reaction (containing no DNA extract) as the last reaction in each PCR. We amplifi ed eluates from both extraction protocols, and results ar
e shown in Fig. 1
. Extraction negatives were previously run in separate reactions and were blank.
We performed maximum likelihood (ML) and ML bootstrap
(MLBP) analyses using RAxML 7.0.4
( 8
) using the GTRGAMMA model. Bayesian phylogenies were estimated using MrBayes v3.2
( 9, 10 )
, applying the GTR+I+G model as selected using jModelt-
est v0.1.1 ( 11
) . Two runs of fi ve million generations each were performed simultaneously, sampling every 200 generations and a 10% burnin. We assessed convergence by visualizing the traces in MrBayes and determining a potential scale reduction factor (PSRF) of ~1.00 for all parameters. Trees were visualized in FigTree v.1.3.1
( http://tree.bio.ed.ac.uk/software/fi gtree
) and the ML tree with MLBP and Bayesian posterior clade probabilities (BPP) is shown
in Fig. 2 .
4 Case Study: Recovery of Ancient Nuclear DNA…
33
Columbina squammata
*
Columbina minuta
91/1.0
Columbina passerina
American ground doves
*
Columbina picui
Metriopelia morenoi
*
Metriopelia ceciliae
Ectopistes migratorius
Passenger pigeon
Patagioenas squamosa
74/0.98
Patagioenas speciosa
84/1.0
Patagioenas oenops
New World pigeons
Patagioenas plumbea
79/0.98
Patagioenas fasciata
Streptopelia chinensis
*
Streptopelia senegalensis
Typical pigeons & do
91/0.98
Streptopelia mayeri
Streptopelia picturata
Streptopelia hypopyrrha
94/1.0
Turtledoves
78/1.0
Streptopelia orientalis
Streptopelia tranquebarica
Streptopelia capicola
91/1.0
Streptopelia decipiens
62/0.91
Streptopelia decaocto
Columbiformes
Columba pulchrichollis
*
Columba palumbus
Columba livia
Old World pigeons
86/1.0
70/0.97
Columba rupestris
64/
Columba guinea
v
90/0.97
0.99
e
Columba arquatrix
s
Macropygia mackinlayi
77/0.98
Macropygia ambionensis
77/0.91
Macropygia phasianella
Cuckoo-doves
*
Reinwardtoena browni
Turacoena manadensis
Leptotila rufaxilla
*
Leptotila cassini
*
Leptotila jamaicensis
Leptotila verreauxi decipens
*
Zenaida auriculata
*
Zenadia aurita
Zenaidine &
Zenaida graysoni
83/0.99
Zenaida asiatica
quail-doves
79/0.99
Zenaida meloda
Geotrygon costaricensis
Geotrygon chiriquensis
86/1.0
*
Geotrygon albifacies
Geotrygon montana
Chalcophaps stephani
*
Chalcophaps indica
*
African wood-doves
Tutur chalcospilos
85/0.99
Oena capensis
& Emerald doves
Turtur brehmeri
Treron vernans
*
Treron calva
Green pigeons
98/1.0
Ptilinops richardsii
96/1.0
Ptilinops pulchellus
Ptilinopus occipitalis
Fruit doves
*
Ptilinopus leclancheri
Ducula rubricera
75/0.97
*
Ducula pistrinaria
Imperial pigeons
Ducula bicolor
97/0.99
Lopholaimus antarcticus
85/1.0
Gymnophaps albertisii
Topknot, mountain & New Zealand pigeons
Hemiphaga novaeseelandiae
Leucosarcia melanoleuca
Petrophassa plumifera
Zebra & Indopacific
Geopelia cuneata
87/0.80
Petrophassa albipennis
ground doves;
Ocyphaps lophotes
Crested, Wonga &
Phaps chalcoptera
84/1.0
Henicophaps albifrons
rock pigeons;
Gallicolumba jobiensis
*
Bronzewings
Gallicolumba beccarii
Goura cristata
Nicobar, pheasant,
Trugon terrestris
crowned & tooth-billed
Otidiphaps nobilis
Caloenas nicobarica
pigeons
Phapitreron leucotis
*
Brown doves
Phapitreron amethystinus
Fig. 2. Molecular phylogeny of
Columbidae
based on a maximum likelihood analysis of available sequences of
FGB intron 7
.
Node support is indicated by maximum likelihood bootstrap (MLBP)/Bayesian posterior probability (BPP). Nodes supported by 100% MLBP and BPP = 1.0 are indicated with a star; nodes with <75% MLBP and BPP <0.95 are unlabeled. Common names are indicated on the
right
, where applicable. Figure is ada
pted from ( 1
).
34
T.L. Fulton
et al.
3. Results
and Discussion
Both the Qiagen kit and silica DNA extraction methods produced amplifi able nuclear DNA fr
om pigeon toe pads (Fig. 1 ). Although
a large elution volume was used in the extraction utilizing the Qiagen kit, a second elution from the same column produced a
band of the same intensity as the other extractions (lane 2, Fig. 1 ),
highlighting the utility of a second elution off the column either at the time of initial extraction or after cold, dry storage (this also works well for silica columns).
Despite the high success rate of DNA recovery and amplifi cation from this sample, several steps could have been taken to increase DNA yield. In subsequent tests with toe pads from other specimens, prolonged exposure to heat, as was applied during the silica extraction, resulted in signifi cant decreases in DNA yield compared to exposure to much shorter periods of warming (48 h). In these experiments, 20 m L of 1 M dithiothreitol (DTT) was added to the extraction buffer to aid in the dissolution of the tissue (B. Letts, personal communication). Following these results, our lab began to routinely include DTT in extractions of bird toe pads using the Qiagen DNeasy extraction kit, and we noted that this step signifi -
cantly reduced the amount of time required to completely dissolve the tissue. When only proteinase K and heat (and EDTA, but this is included primarily for bone decalcifi cation) are applied to dissolve tissue, this process can take over a week; with buffers including detergents to disrupt tissue (as in the Qiagen tissue lysis buffer), dissolution can still take several days. We now routinely perform toe pad extractions by rinsing the intact toe pad in 0.5 M EDTA to wash away inhibitors, and then follow with the Qiagen DNeasy tissue extraction protocol, modifi ed slightly by (1) including 20 m L
of 1 M DTT with the initial tissue lysis step, (2) very gently shaking the samples in a 50°C oven for 48 h, and (3) adding an extra 20 m L
of proteinase K solution after 24 h. We prefer warming in an incubator rather than a heat block, as the heat is more evenly distributed in the incubator. In our experience, most toe pad and desiccated tissue samples are dissolved after the fi rst 24 h and all are dissolved after 48 h. The procedure then proceeds as per the manufacturer’s protocol, although the elution volume is generally reduced to 50 m L, since the eluate can be diluted afterward. This procedure has yielded viable mtDNA (nuclear DNA not tested) from over 90% of pigeon toe pad specimens processed in our lab, including a specimen that was lacquered and several that were on display and exposed to light for many years.
The exceptional preservation of this specimen led to the recovery of a 445 bp sequence of
FGB intron 7
; the fi rst nuclear sequence 4 Case Study: Recovery of Ancient Nuclear DNA…
35
amplifi ed from the extinct passenger pigeon
( 1
) . Maximum likelihood and Bayesian phylogenetic analyses indicate strong support for a sister relationship between the passenger pigeon and other New World
pigeons (Fig. 2 , adapted fr
om
( 1
) ), confi rming a single origin for pigeons in the New World. Phylogenetic analyses and sequence acquisition methods are described in detail in
( 1 )
. In conclusion, we illustrate the possibility of obtaining nuclear DNA from well-preserved historic toe pad specimens from multiple DNA extraction methods to uncover information about an extinct species.
Acknowledgments
We thank Clemency Fisher and the National Museums Liverpool for providing
Ectopistes
material for analysis. Funding for this research was provided by the Pennsylvania State University to BS
and an Undergraduate Discovery Grant to SMW.
References
1. Fulton TL, Wagner SW, Fisher C, Shapiro B (in
Cooper A (2002) Flight of the dodo. Science
press) Nuclear DNA from the extinct Passenger
295:1683
Pigeon (
Ectopistes migratorius
) confi rms its phylo-7. Johnson KP, Clayton DH, Dumbacher JP, genetic placement within Columbinae. Ann Anat
Fleischer RC (2010) The fl ight of the passen—