Regenesis (21 page)
Authors: George M. Church
Cancer-seeking stealth bacteria and humanized mammals for monoclonal antibody production show that genomic engineering can improve human health. Still, these two technologies are mopping-up operations, interventions employed only after a disease occurs. A better application of synthetic genomics would be to prevent diseases from ever occurring by altering the human genome, directly reengineering ourselves to higher levels of health, hardiness, and disease resistance.
If that sounds heretical or insane, consider that human beings have been doing something like this for a long time, even with no knowledge of genetics. When people try to select a mate possessing the best possible qualitiesâstrength, health, stamina, good looksâfor the purpose of producing healthy offspring, the effect, if and when achieved, would result in the creation of a new genome, one whose genes yielded the sought-after physical traits. In more recent history, when women seeking to give birth to “genius” babies applied to the now defunct Repository for Germinal Choice (popularly known as the Nobel Prize sperm bank) for insemination with what they thought was genius-level sperm cells, what they were trying to do was to incorporate high intelligence genes into their children's genomes. (Whether this actually works is another matter.)
So it's not as if we don't already try to selectively alter, improve, and beef-up human genomes. Synthetic genomics attempts to do the same thing systematically while drawing on a detailed scientific understanding of genetics, as well as huge experimental and informational databases pertaining to any proposed genomic alteration.
One of the greatest human health innovations of all time would be to make ourselves multivirus resistantârender ourselves immune to all viruses, known or unknown, whether currently existing or waiting in the wings to come off the evolutionary assembly line in the future. This means that there would be no more influenza pandemics, no more common
colds, no more HIV-AIDS, no more herpes, hepatitis, or polio. And absolutely no more rabies.
We have already seen how we could make human beings multivirus-resistant by mirror-flipping every chiral molecule in our bodies. We could also achieve multivirus resistance by changing our genetic code.
And here we are, at the climax to “the greatest story ever,” the story of the genome. For the genome is written in code, a code that was established by nature and has remained unchanged for billions of years. While countless individual genomes have come and gone during that time, the code on which all of them were, and still are, based has remained unchanged for eons. But now, after all those billions of years, we propose . . . to change it.
Yes, this sounds like hubris all right, not to mention “playing God.” It's almost as if we're proposing to reposition the stars in the heavens, or to rewrite the periodic table of the elements.
In reality, there's nothing blasphemous about the prospect of changing the genetic code to improve human health and prevent illness and death from viral infection. The process is probably easier than changing ourselves into mirror people. There is only one (hard) way to change ourselves into mirror humans, but there are a vast number of ways of changing our genetic code.
We've seen that viruses are able to replicate themselves inside our cells because they use the same genetic code, or speak the same genetic language, as our own cells. One way to defeat the virus's ability to replicate, then, would be to throw a monkey wrench into this chain of events by altering the genetic code of the host cell, as well as that of the cellular machinery that translates the virus's codons, so that the virus's genome becomes unreadable by the host cell and its translation apparatus. More specifically, we're going to alter some of the codons of the host cell's genome, as well as some codons within the cell's translation apparatus, so that these cellular mechanisms no longer recognize the codons of the virus's genome. Instead, the host cell's translation mechanisms will trip up on the virus's codons and effectively come to a halt. At that point, the virus is stopped in its tracks. The viral genetic message that formerly made perfect molecular sense to the host cell now amounts to nonsense, and as far
as the host cell is concerned, the viral invader's genome is incomprehensible molecular babble. The upshot: multivirus resistanceâimmunity to all viruses.
Making changes in our genetic code is not a matter to be taken lightly. The process would start with experiments on bacteria and proceed to ever more complex genomes to mammals before ever trying the procedure on human beings. So we will proceed cautiously, and with due regard to safety. We are working this out initially for industrial microbes like
E. coli
, which are especially susceptible to contamination with viruses (phages), a susceptibility that endangers production vats of millions of liters of pharmaceuticals or chemicals. Such a contamination event occurred with hamster cells at Genzyme in Boston in 2009âand it probably happens more often than we know, since companies don't like to publicize their mistakes.
So, how do we change the genetic code? By removing a key part of the host machinery that the virus needs to replicate itself. But doing that and nothing else, however, would also make the
cell
unable to replicate itself. You can't remove a key part of the host machinery that the virus needs to replicate itself until you have removed every aspect of the cell's genome whose replication also depends on that part. In the following section I'll illustrate what this means with an example that has nothing to do with the genetic code but nevertheless exemplifies the principle stated above.
Once upon a time a pawn shop chain called Buy ân' Cell installed indestructible metal roll-down night gates and equipped them with heavy-duty combination locks for each store. To keep things simple for the employees who opened the shops in the morning, management left the locks at the factory setting of 123456789. But a local gang, aptly called the Striking Virals, wanted to break into these shops to enrich themselves and maybe become even bigger and better and more powerful than before, and go on to raid even more shops.
One night, gang members went to each store and tried the combinations 000000000 and then 123456789. The second number worked perfectly, and
the gang looted each and every shop. The next morning management changed all of the locks to 223456789 but didn't manage to reach all of the employees with the new number. Those stores didn't open. This illustrates the all-important metaphysical principle that you can't change the combination that the employees need to open the store unless you have informed every store employee who depends on knowing that combination in order to open the shop.
The next night, the Virals tried the old combination without success but soon came up with the new combination, since it was only one mutation away from the original. The Buy ân' Cell staff responded the next day by changing the combination to 892220611 and giving the new number to all employees. Later that night, after a few minutes of trying different numbers, a genius-level member of the gang realized that even going at the rate of one try per second, nonstop, it would take them half a billion seconds (fifteen years) to try even half of all the possible combinations. So they gave up and retired from the scene, but not before covering all the metal gates with graffiti copies of Napoleon's famous epigram, “Glory is fleeting but obscurity is forever.”
So how does this stirring tale relate to the process of changing the genetic code?
We start with transfer RNA (tRNA)âfolded molecules that transport amino acids from the cytoplasm of a cell to a ribosome that strings together amino acids into proteins. Each tRNA molecule is essential for the life of the host, unless you can get rid of every instance of the codon that needs that particular tRNA to transport an amino acid to the ribosome. To get rid of those codons you'd need to swap each of them with a so-called synonymous codon that uses a different tRNA but translates to the same amino acid. Or, in terms of the parable, to do that you'd need to swap that combination (codon) with a so-called synonymous combination (codon) that uses different numbers (codon letters) but performs the same function (prompting a tRNA to transport the same amino acid to the ribosome). (A synonymous codon is one whose different nucleotides encode the same amino acid; for example, both AGA and CGG encode the amino acid arginine.)
Our first goal in the lab was to remove the gene for the protein called “release factor 1” (nicknamed RF1). This protein is required for
E. coli
ribosomes to stop adding amino acids at the end of making any of 322 different proteins that terminate in the stop codon UAG. RF1 binds to the mRNA at the UAG and causes the ribosome to stop. We couldn't delete RF1 until we changed all 322 instances of UAG to the synonymous UAA codon (which is handled by a different protein known as RF2). When an invading virus comes along, it needs the RF1 to use the UAG codon, but RF1 isn't there anymore! The ribosome will still manage to function, but badly. Nevertheless, probably two or more codon changes will be needed to defeat all viruses.
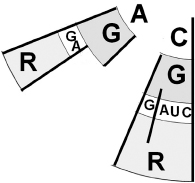
Figure 5.2
A small segment of the genetic code (from
Figure 3.2
) showing all the six synonymous codons for the amino acid arginine (R): AGA, AGG, CGC, CGU, CGA, and CGG.
When we began these experiments we weren't sure that these so-called synonymous codons would all be truly equivalent, because living things tend to use their parts in many gloriously complex and overlapping ways. Ways that would be considered “spaghetti code” if a programmer ever tried such undisciplined and tortuous protocols. However, we did successfully make all of the 322 changes. For our synthetic purposes (not necessarily for all of the natural purposes of the cell), these changes are innocuous and hence we can take the next steps toward multivirus resistance.
How did we manage to make so many changes? The answer lies in the process and instrument called MAGE, introduced in
Chapter 3
. That process allows us to make many changes simultaneously and in parallel.
Next we target the tRNA that recognizes the AGA and AGG codons, normally translating these to the amino acid arginine (R). In
Figure 5.2
we see the six synonymous codons for arginine. If every one of the AGA and AGG codons is changed to CGA, CGG, CGC, or CGU, all of which
also
code for arginine, then we can remove the gene for the tRNA that
binds, recognizes, and translates the AGA and AGG codons (otherwise removing them would be lethal to the cell). If that tRNA is missing, the ribosome will stall at each AGA and AGG and make defective versions of essential cell proteins. Even if other tRNAs jump in and prevent stalling, the number of incorrect amino acids would be disastrous to the cell. Such genome-wide reassignment of codons can be extended to still other codons and then the corresponding tRNAs can be deleted. For example, leucine (L), like arginine, has six synonymous codons.
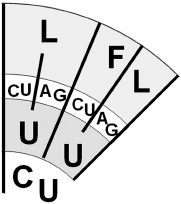
Figure 5.3
A small segment of the genetic code (from
Figure 3.2
) showing all six synonymous codons for the amino acid leucine (L): CUC, CUU, UUC, UUU, UUA, and UUG.
With regard to the piece of genetic code in
Figure 5.3
, we can now free up (make nonessential) one of the three tRNA molecules for leucine by moving all of the CUC and CUU codons to CUA or CUG. At that point, the host has freed up (eliminated all uses of) five of the sixty-four codons (UAG, AGA, AGG, CUC, CUU) and we have deleted three genes that are normally essential to translation and hence are essential for life. This means that invading viruses not only can't use the cell machinery but they don't even have a chance of mutating to use it.
As a concrete, real-life example of this, one of the smallest functional peptides in all of biology has the five amino-acid sequence MRLFV (methionine-arginine-leucine-phenylalaline-valine, which provides resistance to the antibiotic erythromycin). If the peptide were initially encoded by the following five triplets,