Regenesis (28 page)
Authors: George M. Church
In 2005 the nine BioFab co-conspirators wrote a paperâa sort of BioFab manifestoâdescribing how we intended to transform biological engineering from a craft into an industry. It appeared in 2006 as a
Scientific
American
cover story: “Engineering Life: Building a FAB for Biology.” Many of the techniques and devices, principles and practices we proposed there are now staples of the second generation of commercial synthetic genomics.
In general, fundamental engineering principles include (1) the use of interoperable parts (e.g., USB ports and plugs, dual inline pins for chips); (2) standards (e.g., maximum variation in properties, stress resistance); (3) hierarchical functional designs (e.g., designing the windshield without worrying about the hubcap design); (4) computer-aided design (CAD); (5) cost-effectiveness (e.g., miniaturization of integrated circuits, assembly lines); (6) physical isolation (e.g., car windows, plastic-coated insulated electric wires); (7) functional isolation (e.g., car keys, FCC regulations on radio emissions from circuits); (8) redundant systems (e.g., seat belts plus roll bars and air bags); (9) ruggedness and safety testing (e.g., crash dummies); (10) user licensing (e.g., driver and car registration); (11) user surveillance (e.g., radar monitoring of individual vehicle speed, stoplight cameras); (12) functional integration (e.g., dashboard clustering of instrumentation); (13) evolution (e.g., field testing and market feedback). Items 6-11 are also relevant to safety and security.
The biological equivalents to the principles and examples mentioned above are (1) interoperable parts (e.g., biobricks; see
Chapter 8
); (2) standards (e.g., protocols for connecting one biological part to the next; see
Chapter 8
); (3) hierarchical functional designs (e.g., designing transcription termination sites levels without worrying about how they start); (4) computer-aided design (CAD) (e.g., caDNAnoâsoftware for the design of three-dimensional DNA origami structures); (5) cost-effectiveness (e.g., choosing organisms that replicate quickly and do not require lots of expensive nutrients, land, energy, or other scarce resources); (6) physical isolation (e.g., sterilizable bioreactors, biosafety level BSL4 labs, and moon suits); (7) functional isolation (e.g., new genetic codes; see
Chapters 3
and
5
); (8) redundant systems (e.g., nutritional constraints plus isolation; item 6); (9) ruggedness and safety testing (e.g., building radiation resistance into organisms, testing genomically modified organisms in biosafety labs before releasing them into the environment); (10) user licensing (e.g., registration
of DNA synthesis machines, chemicals, and lab personnel); (11) user surveillance (e.g., requiring commercial DNA synthesis firms to check orders against known pathogen sequences); (12) functional integration (e.g., codesigning photo-bioreactors and cyanobacterial genomes); (13) evolution (e.g., MAGE, sensor selectors). (For items 6â13, see the Epilogue.)
Item 13, evolution, differs vastly between biological and other types of engineering. In nonbiological engineering, huge resources go into developing a new product, and so there is rarely more than one or a few new products in a given niche per company, and only a few companies worldwide operating in those niches. In biological engineering, however, researchers can intelligently design billions of genomes and, given clever selection (see
Chapter 3
), one or a few winners will emerge from a labscale version of Darwinian survival of the fittest. And each year we get faster and better at laboratory versions of evolution.
Just as the fundamental principles of engineering apply to biology, so basic business principles are being applied and adapted to the field of genomic engineering. The process of extending engineering principles to biology and applying business principles to genome engineering were logical and indeed inevitable developments. But at the beginning, these ideas were so counterintuitive that it took forty years from the dawn of molecular biology to the dawn of molecular biology engineering, and about twenty years to adjust to the business challenges and even to the philosophical and policy components of the new business models. Some business practices that are taken for granted in mechanical, chemical, civil, and aeronautical engineering took on strange twists when applied to human beings and form a type of “biological exceptionalism.” By exceptionalism, I mean that an invention that is useful and nonobvious would be embraced if made from chemicals, but not if made from cells. What people debating the topic of “patenting life” sometimes forget is that the alternative to patenting biological inventions is a burgeoning of trade secrets. Truly creative or serendipitous breakthroughs will get hidden, possibly for years, until rediscovered
independently. A patent is a limited monopoly to coax such secrets out into the open. Seldom have such monopolies impeded research. Occasionally there are rumors of intimidationâfor example, labs making homebrew enzymes (Taq-polymerase, owned by Roche) for do-it-yourself gene amplification. The scariest examples of monopoly are tools for targeted DNA recombination (Cre-lox, owned by Dupont) and diagnostic breast cancer genetic diagnostics
(BRCA1
and
BRCA2
, owned by Myriad Genetics).These patent monopolies probably don't prevent research, but they may be perceived as preventing or reducing profits for new inventions building on the older ones.
The term “synthetic biology” originated in 1912 with Stéphane Leduc's work “La biologie synthétique, étude de biophysique,” but has been embraced since 2004 to provide an intentional new feel to the enterprise, to distance ourselves from previous fields like genetic engineering, and to encourage a new rigor around the dozen or so fundamental engineering principles listed above. But the field soon splintered into six different camps: (1) recombinant DNA applied to metabolic engineering (Keasling), (2) bio-inspired and bio-mimetic (Steve Benner and Bob Langer), (3) engineering-inspired biobricks (Tom Knight), (4) evolution (Arnold, Ellington, Church), (5) instrumentation (the Wyss Institute and various companies), and (6) genome engineering (JCVI, HMS).
It makes a huge difference which camp you bet on. Critics might make the following objections: (1) Old-fashioned single gene bashing is low throughput. (2) Mimics don't take full advantage of real biology. (3) The ideas of modularity and biocircuits are naive, context-dependent, and not robust. (4) Historically evolution is not fast (relative to quarterly reports). (5) While the markets for bioproducts may be huge, the market for machines to improve production may be tiny. And as costs plummet, how do you expand this tiny market? (6) Why synthesize a whole genome if you only need to change a few bits?
Entrepreneurship is not for the faint of heart, and this is just as true for the synthetic genomics business as it is for any other. Companies, like technologies, tend to come and go. Indeed, the reading and writing of DNA probably constitute the fasting moving technology freefall in history (
Figure 7.1
). There are multiple pathways ahead, and so companies can merge with other companies. They can get acquired by other companies, and their technology and people can be split up in various ways. With all that in mind, we turn to a capsule history of genomic engineering as a business, a terrible and awesome saga, complete with vignettes of major players, personal asides, and even pithy apothegms.
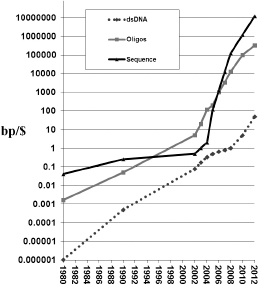
Figure 7.1
Exponential technologies. Gordon Moore's law for the density of transistors in large-scale integrated circuits has accurately held at a spectacular exponential 1.5-fold improvement per year for the cost of various aspects of electronics, disk drives, telecommunications, and applications since the mid-1960s (or perhaps all the way back to semaphores in the 1790s). Similarly, DNA reading and writing technologies were on the same (1.5-fold per year) curve until 2005, when so-called next generation technologies started to arrive and the exponential shifted to an almost tenfold increase per year. While in principle you can automate anything, automation alone doesn't guarantee rapid cost changes. The new (tenfold per year) curves required a radical shift in methods from the use of columns through which DNA flows to the use of flat glass on which multiplex immobilized DNA reactions can occur and hence be downsized in volume and scaled up in numbers. We now can multiplex billions of reading reactions in one flow cell and 30 million oligos synthesized on one wafer (e.g., using Agilent ink-jet process).
You might think that the best way to obtain government funding is to lobby Congress or form a blue ribbon advisory committee at the august National Academy of Sciences or form a grassroots movement that gathers signatures. But then you would miss the swiftest and most powerful motivator of all: fear. In 1987, 1995, and 1998 three companies, working independently, received billions of dollars from the United States and the world for a previously unknown backwater of biomedical research: genome sequencing.
In the spring of 1987 Wally Gilbert, onetime CEO of the Cambridge biotechnology firm Biogen, decided that the time was ripe for a company to prospect and lay claim to as much of the human genome as possible, and swiftly. He used his Biogen golden parachute to recruit the pack that would later found the Human Genome Project (HGP)âLee Hood, Sydney Brenner, Eric Lander, Charles Cantor, and myselfâand announced the formation of a company with the working name Genome Corporation, for the purpose of sequencing the human genome and copyrighting and selling the data. (The name Terabase was used internally as a play on terabase meaning a trillion DNA bases and terrabase meaning a base for world domination.)
But the stock market crash of October 1987 made the capitalization of the Genome Corporation impossible, and so the company died on the operating table, as it were. Nevertheless, the public reaction to our plan to
sell human genomic information
(!) was speedy and alarmist: we do not want any company “owning” our genes, our priceless genetic heritage, our children's future. Congress appropriated $3 billion to NIH over the next fifteen years to sequence the human genome (a buck a base), and voted to make the information free to all humanity before any company could claim it. This was framed as a new budget line item to avoid criticism from entrenched NIH grantees that mediocre “big science” (huge academic genome centers) would steal from ongoing “small but great science” (the science of the small grantees, naturally).
The second fear genie was unleashed in 1992, ironically, when NIH lawyers insisted on patenting bits of human source code that Craig Venter's team had sequenced from RNA molecules. This got enough negative attention that a major investor, Health Care Investment Corporation, coughed up $70 million to form Venter's nonprofit TIGR (The Institute for Genomic Research) and the for-profit Human Genome Sciences (HGS). The genes TIGR sequenced and patented would be licensed to HGS for commercialization. That arrangement poured fuel on the flames, and the public launched a supplementary effort, the Merck Gene Index Project (MGIP), on top of the already huge Human Genome Project to directly compete with HGS. This supplementary effort involved companies that normally would not work together, but in this case were less threatened by each other than by a single company (HGS) owning most of the human genome. The reactionary tactic worked (sort of). During the ensuing fool's gold rush, the patents filed by TIGR did not generally add sufficient value to get far enough away from unpatentable “products of nature.” Nevertheless the juggernaut of sequencing mRNA molecules would continue, especially once next-generation sequencing made it a viable way to quantitate RNA levels.
The third and largest genie appeared in 1998 when the PE (Perkin-Elmer) Corporation, later renamed Applera, created Celera. This was the private firm, successor to TIGR, where Venter used the shotgun sequencing method (initiated by Roger Staden in 1979 and applied to bacteria in 1995 at TIGR) to essentially speed-sequence the human genome. (Francis Collins, director of the competing public Human Genome Project, and no big fan of Venter's, said that shotgun sequencing would produce “the Cliffs Notes or the Mad magazine version” of the genome.) Venter's plan at Celera was to patent certain gene sequences and license them to commercial firms for a fee, while at the same time publishing unpatented sequences on its web page.
Coincidentally, Celera exhibited the newest model of Applera's sequencer (at the time known as the Applied Biosystems sequencer). This in turn drove sales of the sequencer to both sides in the human genome Olympics.
So who won? As Nietzsche once said, “All and none.”
In June 2000 Venter and Collins, standing on either side of Bill Clinton, announced the completion of the draft human genome to the public. John Sulston, head of the small British genome project, said in his book about the HGP,
The Common Thread
, “The date of the announcement, 26 June, was picked because it happened to be free in both Bill Clinton's and Tony Blair's diaries. [There was a simultaneous UK announcement on the same date.] It was not clear that the Human Genome Project had quite got to its magic 90 percent mark by then, and Celera's data were invisible but known to be thin, so nobody was really ready to announce; but it became politically inescapable to do so. We just put together what we did have and wrapped it up in a nice way, and said it was done. . . . Yes, we were just a bunch of phonies!”