Power, Sex, Suicide: Mitochondria and the Meaning of Life (11 page)
Read Power, Sex, Suicide: Mitochondria and the Meaning of Life Online
Authors: Nick Lane
Tags: #Science, #General
The Hydrogen Hypothesis
The quest to find the progenitor of the eukaryotic cell has run into dire straits. The idea that there might have been a primitive intermediate, a missing link with a nucleus but no mitochondria, has not been rigorously disproved, but looks more and more unlikely. Every promising example has turned out
not
to be a missing link at all, but rather to have adapted to a simpler lifestyle at a later date. The ancestors of all these apparently primitive groups
did
possess mitochondria, and their descendents eventually lost them while adapting to new niches, often as parasites. It seems possible to
be
a eukaryote without having mitochondria—there are a thousand such species among the protozoa—but it does not seem possible to be a eukaryote without once having
had
mitochondria, deep in the past. If the only way to be a eukaryotic cell is via the possession of mitochondria, then it might be that the eukaryotic cell itself was originally crafted from a symbiosis between the bacterial ancestors of the mitochondria and their host cells.
If the eukaryotic cell was born of a merger between two types of cell, the question becomes more pressing—what types of cell? According to the textbook view, the host cell was a primitive eukaryotic cell, without mitochondria, but this obviously can’t be true if there never was a primitive eukaryotic cell that lacked mitochondria. In her endosymbiosis theory, Lynn Margulis had in fact proposed a union between two different types of bacteria, and her hypothesis looked set for a return to prominence after the demise of the missing link. Even so, Margulis and everyone else were thinking along the same lines—the host, they imagined, must have relied on fermentation to produce its energy, in the same way that yeasts do today, and the advantage that the mitochondria brought with them was an ability to deal with oxygen, giving their hosts a more efficient way of generating energy. The exact identity of the host could potentially be traced by comparing the gene sequences of modern eukaryotes with various groups of bacteria and archaea—and modern sequencing technology was just beginning to make that possible. But, as we have just seen, the apparent answer came as another shock: the genes of eukaryotic cells seem to be related most closely to
methanogens
, those obscure methane-producing archaea that live in swamps and intestines.
Methanogens! This answer is an enigma. In
Chapter 1
, we noted that the methanogens live by reacting hydrogen gas with carbon dioxide, and evanescing methane gas as a waste product. Free hydrogen gas only exists in the absence of oxygen, so the methanogens are restricted to
anoxic
environments—any marginal places where oxygen is excluded. It’s actually worse than that. Methanogens can tolerate some oxygen in their surroundings, just as we can survive underwater for a short time by holding our breath. The trouble is that methanogens can’t generate any energy in these circumstances—they have to ‘hold their breath’ until they get back to their preferred anoxic surroundings, because the processes by which they generate their energy can
only
work in the strict absence of oxygen. So if the host cell really was a methanogen, this raises a serious question about the nature of the symbiosis—why on earth would a methanogen form a relationship with any kind of bacteria that relied upon oxygen to live? Today, modern mitochondria certainly depend on oxygen, and if it was ever thus, neither party could make a living in the land of the other. This is a serious paradox and did not seem possible to reconcile in conventional terms.
Then in 1998, Bill Martin, whom we met in
Chapter 1
, stepped into the frame, presenting a radical hypothesis in
Nature
with his long-term collaborator Miklós Müller, from the Rockefeller University in New York. They called their theory the ‘hydrogen hypothesis’, and as the name implies it has little to do with oxygen and much to do with hydrogen. The key, said Martin and Müller, is that hydrogen gas can be generated as a waste product by some strange mitochondria-like organelles called
hydrogenosomes
. These are found mostly among primitive single-celled eukaryotes, including parasites such as
Trichomonas vaginalis
, one of the discredited ‘archezoa’. Like mitochondria, hydrogenosomes are responsible for energy generation, but they do this in bizarre fashion by releasing hydrogen gas into their surroundings.
For a long time the evolutionary origin of hydrogenosomes was shrouded in mystery, but a number of structural similarities prompted Müller and others, notably Martin Embley and colleagues at the Natural History Museum in London, to propose that hydrogenosomes are actually related to mitochondria—they share a common ancestor. This was difficult to prove as most hydrogenosomes have lost their entire genome, but it is now established with some certainty.
1
In other words, whatever bacteria entered into a symbiotic
relationship in the first eukaryotic cell, its descendents numbered among them both mitochondria
and
hydrogenosomes. Presumably, said Martin—and this is the crux of the dilemma faced today—the original bacterial ancestor of the mitochondria
and
hydrogenosomes was able to carry out the metabolic functions of both. If so, then it must have been a versatile bacterium, capable of oxygen respiration as well as hydrogen production. We’ll return to this question in a moment. For now, lets simply note that the ‘hydrogen hypothesis’ of Martin and Müller argues that it was the hydrogen metabolism of this common ancestor, not its oxygen metabolism, which gave the first eukaryote its evolutionary edge.
Martin and Müller were struck by the fact that eukaryotes containing hydrogenosomes sometimes play host to a number of tiny methanogens, which have gained entry to the cell and live happily inside. The methanogens align themselves with the hydrogenosomes, almost as if feeding (
Figure 3
). Martin and Müller realized that this was exactly what they were doing—the two entities live together in a kind of metabolic wedlock. Methanogens are unique in that they can generate all the organic compounds they need, as well as all their energy, from nothing more than carbon dioxide and hydrogen. They do this by attaching hydrogen atoms (H) onto carbon dioxide (CO
2
) to produce the basic building blocks needed to make carbohydrates like glucose (C
6
H
12
O
6
), and from these they can construct the entire repertoire of nucleic acids, proteins, and lipids. They also use hydrogen and carbon dioxide to generate energy, releasing methane in the process.
While methanogens are uniquely resourceful in their metabolic powers, they nonetheless face a serious obstacle, and we have already noted the reason in
Chapter 1
. The trouble is that, while carbon dioxide is plentiful, hydrogen is hard to come by in any environment containing oxygen, as hydrogen and oxygen react together to form water. From the point of view of a methanogen, then, anything that provides a little hydrogen is a blessing. Hydrogenosomes are a double boon, because they release both hydrogen gas and carbon dioxide, the very substances that methanogens crave, in the process of generating their own energy. Even more importantly, they don’t need oxygen to do this—quite the contrary, they prefer to avoid oxygen—and so they function in the very low-oxygen conditions required by methanogens. No wonder the methanogens suckle up to hydrogenosomes like greedy piglets! The insight of Martin and Müller was to appreciate that this kind of intimate metabolic union might have been the basis of the original eukaryotic merger.
Bill Martin argues that the hydrogenosomes and the mitochondria stand at opposite ends of a little-known spectrum. Rather surprisingly, to anyone who is most familiar with textbook mitochondria, many simple single-celled eukaryotes have mitochondria that operate in the absence of oxygen. Instead of using oxygen to burn up food, these ‘anaerobic’ mitochondria use other simple compounds like nitrate or nitrite. In most other respects, they operate in a very similar fashion to our own mitochondria, and are unquestionably related. So the spectrum stretches from aerobic mitochondria like our own, which are dependent on oxygen, through ‘anaerobic’ mitochondria, which prefer to use other molecules like nitrates, to the hydrogenosomes, which work rather differently but are still related. The existence of such a spectrum focuses attention on the identity of the ancestor that eventually gave rise to the entire spectrum. What, asks Martin, might this common ancestor have looked like?
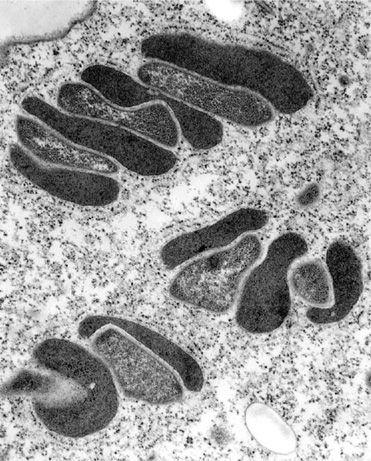
3
The image shows methanogens (light grey) and hydrogenosomes (dark grey). All are living inside the cytoplasm of a much larger eukaryotic cell, specifically the marine ciliate
Plagiopyla frontata
. According to the hydrogen hypothesis, such a close metabolic relationship between methanogens (which need hydrogen to live) and hydrogen-producing bacteria (the ancestor of the mitochondria as well as hydrogenosomes) may have ultimately given rise to the eukaryotic cell itself: the methanogens became larger, to physically engulf the hydrogen-producing bacteria.
This question has profound significance for the origin of the eukaryotes, and so for all complex life on earth or anywhere else in the universe. The common ancestor could have taken one of two forms. It could have been a sophisticated bacterium with a large bag of metabolic tricks, which were later distributed to its descendents, as they adapted to their own particular niches. If that were the case, then the descendents could be said to have ‘devolved’, rather than ‘evolved’, for they became simpler and more streamlined as they grew specialized. The second possibility is that the common ancestor was a simple oxygen-respiring bacterium, perhaps the free-living ancestor of
Rickettsia
we discussed in the previous chapter. If that were the case, then its descendents must have become more diverse over evolution—they ‘evolved’ rather than ‘devolved’. The two possibilities generate specific predictions. In the first case, if the ancestral bacterium was metabolically sophisticated, then it was in a position to hand down specialized genes directly to its ancestors, such as those for hydrogen production. Any eukaryotes adapting to hydrogen production could have inherited its genes from this common ancestor, regardless of how diverse they were to become later. Hydrogenosomes are found in diverse groups of eukaryotes. If they inherited their hydrogen-producing genes from the same ancestor, then these genes should be closely related to each other, regardless of how diverse their host cells became later. On the other hand, if all the diverse groups had originally inherited simple, oxygen-respiring mitochondria, they had to invent all the different forms of anaerobic metabolism independently, whenever they happened to adapt to a low-oxygen environment. In the case of the hydrogenosomes, the hydrogen-producing genes would necessarily have evolved independently in each case (or transferred randomly by lateral gene transfer), and so their evolutionary history would be just as varied as that of their host cells.
These possibilities give a plain choice. If the ancestor was metabolically sophisticated, then all the hydrogen-producing genes should be related, or at least
could
be related. On the other hand, if it was metabolically simple, then all these genes should be unrelated. So which is it? The answer is as yet unproved, but with a few exceptions, most evidence seems to favour the former proposition. Several studies published in the first years of the millennium attest to a single origin for at least a few genes in the anaerobic mitochondria and hydrogenosomes, as predicted by the hydrogen hypothesis. For example, the enzyme used by hydrogenosomes to generate hydrogen gas (the pyruvate: ferredoxin oxidoreductase, or PFOR), was almost certainly inherited from a common ancestor. Likewise the membrane pump that transports ATP out of
both mitochondria and hydrogenosomes seems to share a similar ancestry; and an enzyme required for the synthesis of a respiratory iron-sulphur protein also appears to derive from a common ancestor. These studies imply that the common ancestor was indeed metabolically versatile and could respire using oxygen or other molecules, or generate hydrogen gas, as the circumstances dictated. Critically, such versatility (which might otherwise sound somewhat hypothetical) does exist today in some groups of α-proteobacteria such as
Rhodobacter
, which might therefore resemble the ancestral mitochondria better than does
Rickettsia
.
If so, why is the
Rickettsia
genome so similar to modern mitochondria? Martin and Müller argue that the parallels between
Rickettsia
and mitochondria derive from two factors. First,
Rickettsia
are α-proteobacteria, so their genes for
aerobic
(oxygen-dependent) respiration should indeed be related to the genes of
aerobic
mitochondria, as well as to those of other free-living oxygen-dependent α-proteobacteria. In other words, the mitochondrial genes are similar to those of
Rickettsia
not because they are necessarily
derived
from
Rickettsia
, but because
Rickettsia
and mitochondria both derived their genes for aerobic respiration from a common ancestor that might have been very different to
Rickettsia
. If that is the case, it begs the question: why, if they derived from a very different ancestor, did they eventually become so similar? This brings us to the second point postulated by Martin and Müller—they did so by
convergent evolution
, as discussed at the beginning of
Part 1
. Both
Rickettsia
and mitochondria share a similar lifestyle and environment: both generate energy by aerobic respiration inside other cells. Their genes are subject to similar selection pressures, which might easily bring about convergent changes in both the spectrum of surviving genes, and in their detailed DNA sequence. If convergence is responsible for the similarities, then the genes of
Rickettsia
should only be similar to the mammalian
oxygen-dependent
mitochondria, and not to the other types of
anaerobic
mitochondria that we have been discussing in the last few pages. If the common ancestor was very different to
Rickettsia
—if it was actually a versatile bacterium like
Rhodobacter
, with a bag of metabolic tricks—we wouldn’t expect to find parallels between
Rickettsia
and these anaerobic mitochondria; and for the most part we do not.